During early animal embryonic development, the parental genomes undergo extensive three-dimensional chromatin restructuring after fertilization. Topologically associating domains (TADs) and compartments rapidly dissolve, leaving chromatin in a highly relaxed state. The gradual reestablishment of higher-order chromatin architecture coincides with zygotic genome activation (ZGA), a highly conserved process observed across all studied early embryos in diverse species including fly, frog, zebrafish, medaka fish, mouse, pig, and human. However, how chromatin architecture is established during and how it interplays with transcription in early mammalian embryos remain elusive.
On August 13, 2025, Professor Xie Wei’s group at the School of Life Sciences, Tsinghua University, published a research article titled “Establishment of chromatin architecture interplays with embryo hypertranscription” in Nature. This study systematically elucidates the regulatory mechanisms underlying the progressive establishment of higher-order chromatin structure in early mouse embryos and uncovers an interplay with a unique “hypertranscription” state, which both shapes and is fostered by the three-dimensional genome organization.
The key architectural protein CTCF remains stably bound to the genome throughout early mouse development. Further analysis identified two types of variable CTCF binding: one showing weak binding at the 1–2 cell stage but significantly increasing after the 8-cell stage, primarily localized at promoters and associated with certain ZGA genes; the other exhibiting strong binding from the 1–8 cell stages but rapidly gone in the inner cell mass (ICM) and later stages, enriched at SINE B2 repetitive elements. The reduction of CTCF binding at B2 sites coincided with increased expression of the transcription factor ADNP during the same period. ADNP competes with CTCF for binding at these B2 sites; conversely, ADNP KO embryos exhibited restored CTCF binding at B2 loci and enhanced chromatin insulation.
In contrast to the stable CTCF binding, another critical architectural complex, cohesin, showed very low binding levels at the 1-cell stage. This correlates with the low translation levels of cohesin components and its loader NIPBL, as well as high expression of the unloader WAPL. The combination of weak loading and strong unloading of cohesion leads to insufficient cohesin-mediated loop extrusion. Increased expression of cohesin and NIPBL coupled with downregulation of WAPL facilitates the gradual restoration of TAD structures.
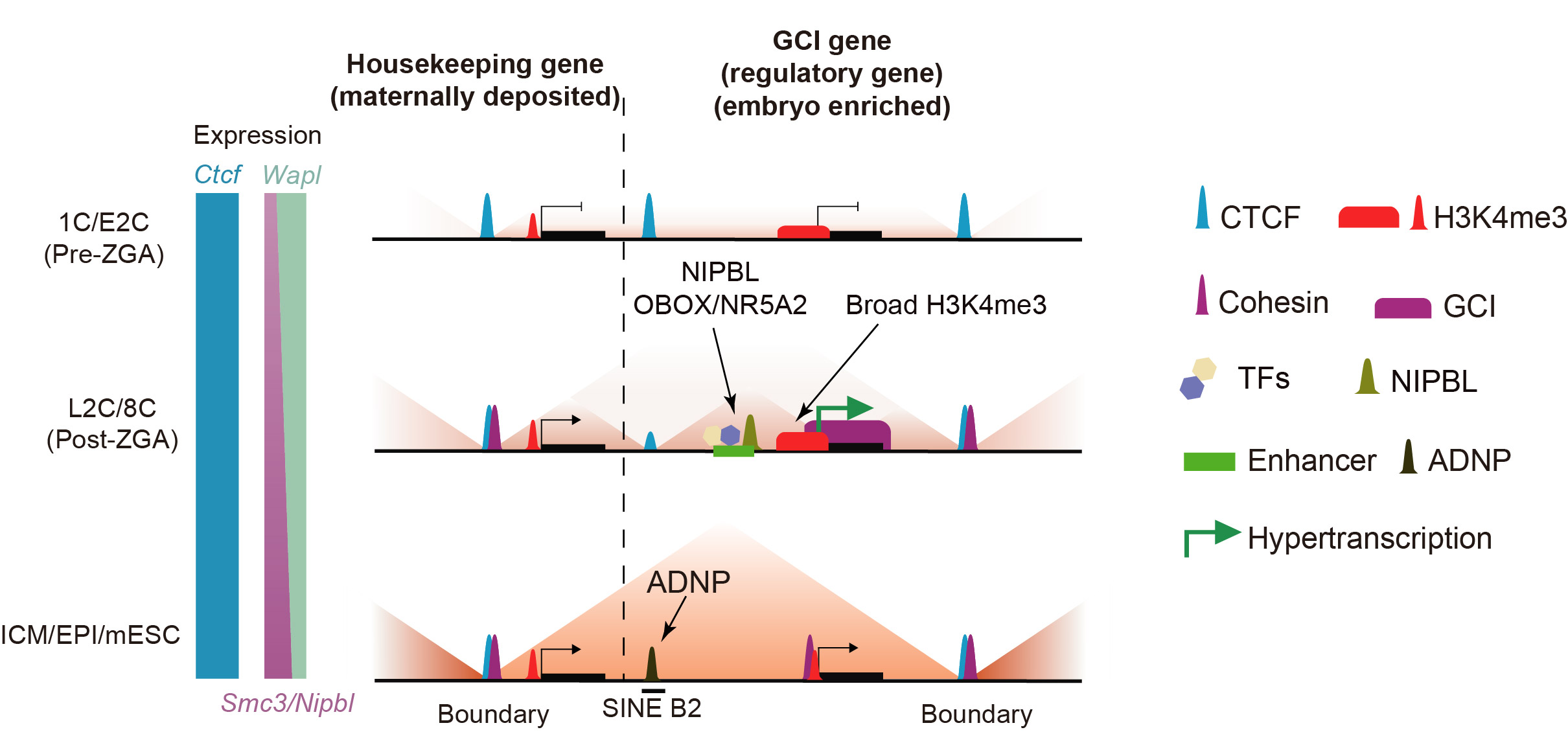
Figure 1. De novo establishment of chromatin architecture and its interplay with hypertranscription in mouse early embryos
Notably, a unique cohesin enrichment pattern termed “genic cohesin islands (GCIs)” is found on actively transcribed genes during the 2–8 cell stages. GCIs are highly specific to the 2–8C stages, and can barely be found in the ICM and later stages. GCIs strongly colocalize with RNA polymerase II (Pol II). Transient inhibition of transcription elongation led to rapid loss of GCIs, while transcription induction generated new GCIs in embryos, indicating that GCIs specifically depend on active transcription. Quantitative analysis of Pol II binding and transcriptome revealed that embryos between the 2–8 cell stages are in a “hypertranscription” state, with transcription levels 6–10 times higher than in differentiating embryonic stem cells and 2–3 times higher than during gastrulation, supporting the model that high densities of elongating RNA polymerase are responsible for the GCI formation.
GCI genes are enriched for key regulatory genes involved in chromatin organization, developmental regulation, and cell fate determination. These genes exhibit broad promoters H3K4me3, and enrich for active enhancers and binding of key TFs and NIPBL in their neighbour regions, providing abundant cohesin loading sites for GCI formation. Previous studies suggest that RNA polymerase can act as a barrier by pushing or hinder it sliding on chromatin. Consistently, GCIs function as transcription-dependent insulators, cooperating with adjacent CTCF sites to form TAD-like interaction domains, without obvious directional preference. Partial cohesin knockdown weakened GCI boundaries and downregulate GCI gene expression, ultimately causing embryonic arrest at the morula stage. Further analyses showed that GCI genes, as key regulators, exhibit low cell-to-cell transcription variability, whereas cohesin KD increases transcriptional heterogeneity among blastomeres of 8C embryos, suggesting that GCIs may help maintain stable expression of critical regulatory genes during embryonic development.
In summary, this study uncovers the interplay between chromatin architecture reestablishment and hypertranscription in early mouse embryos: following ZGA, embryos jump-start their transcription; high-density Pol II stalls cohesin sliding within gene bodies to form GCIs and form contact domains with nearby chromatin insulators as CTCF. This may lead to a positive feedback loop that further fuels hypertranscription and stabilize the transcription variance. This work provides new insights into the interplay between transcription and chromatin architecture, when they are established for the first time.
Professor Wei Xie from School of Life Science at Tsinghua University is the corresponding author of this work. Guang Yu, Kai Xu and Weikun Xia (postdoc fellow) from CLS program at Tsinghua University and Ke Zhang (Ph.D student) from Tsinghua University are the co-first authors of this work. Dr. Qianhua Xu, Dr. Lijia Li, Ling Liu, Dr. Zili Lin, Dr. Bofeng Liu and Dr. Zhenhai Du from Wei Xie’s lab have also made important contributions to this work. Dr. Huili Wang from Jiangsu Academy of Agricultural Sciences, Dr. Haiqiang dai and Dr. Chao Wang from Shanghai Institute of Biochemistry and Cell Biology (Chinese Academy of Sciences), Dr. Hongtao Yu and Dr. Zhubing Shi from Westlake University have provided experimental guidance and essential research materials. This work was supported by the National Key R&D Program of China, the National Natural Science Foundation of China, the Postdoctoral Science Foundation of China and the Tsinghua-Peking Center for Life Sciences. Wei Xie is a recipient of an HHMI International Research Scholar award and a New Cornerstone Investigator.
References:
1. Wang, W., Gao, R., Yang, D., Ma, M., Zang, R., Wang, X., ... & Gao, Y. (2024). ADNP modulates SINE B2-derived CTCF-binding sites during blastocyst formation in mice. Genes & Development, 38(3-4), 168-188.
2. Busslinger, G. A., Stocsits, R. R., Van Der Lelij, P., Axelsson, E., Tedeschi, A., Galjart, N., & Peters, J. M. (2017). Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl. Nature, 544(7651), 503-507.
3. Banigan, E. J., Tang, W., van den Berg, A. A., Stocsits, R. R., Wutz, G., Brandão, H. B., ... & Mirny, L. A. (2023). Transcription shapes 3D chromatin organization by interacting with loop extrusion. Proceedings of the National Academy of Sciences, 120(11), e2210480120.
4. Heinz, S., Texari, L., Hayes, M. G., Urbanowski, M., Chang, M. W., Givarkes, N., ... & Benner, C. (2018). Transcription elongation can affect genome 3D structure. Cell, 174(6), 1522-1536.
Link:https://www.nature.com/articles/s41586-025-09400-5
Xie Lab website:www.xielab.org.cn
